Vectorisation
Last updated on 2024-07-25 | Edit this page
Overview
Questions
- How can I avoid looping over each element of large data arrays?
Objectives
- Use surface land fraction data to mask the land or ocean.
- Use the vectorised operations available in the
numpylibrary to avoid looping over array elements.
A useful addition to our
plot_precipitation_climatology.py script would be the
option to apply a land or ocean mask. To do this, we need to use the
land area fraction file.
PYTHON
import numpy as np
import xarray as xr
accesscm2_sftlf_file = "data/sftlf_fx_ACCESS-CM2_historical_r1i1p1f1_gn.nc"
ds = xr.open_dataset(accesscm2_sftlf_file)
sftlf = ds["sftlf"]
print(sftlf)OUTPUT
<xarray.DataArray 'sftlf' (lat: 144, lon: 192)>
array([[100., 100., 100., ..., 100., 100., 100.],
[100., 100., 100., ..., 100., 100., 100.],
[100., 100., 100., ..., 100., 100., 100.],
...,
[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.]], dtype=float32)
Coordinates:
* lat (lat) float64 -89.38 -88.12 -86.88 -85.62 ... 86.88 88.12 89.38
* lon (lon) float64 0.9375 2.812 4.688 6.562 ... 353.4 355.3 357.2 359.1
Attributes:
standard_name: land_area_fraction
long_name: Percentage of the grid cell occupied by land (including...
comment: Percentage of horizontal area occupied by land.
units: %
original_units: 1
history: 2019-11-09T02:47:20Z altered by CMOR: Converted units fr...
cell_methods: area: mean
cell_measures: area: areacellaThe data in a sftlf file assigns each grid cell a percentage value between 0% (no land) to 100% (all land).
OUTPUT
100.0
0.0To apply a mask to our plot, the value of all the data points we’d
like to mask needs to be set to np.nan. The most obvious
solution to applying a land mask, for example, might therefore be to
loop over each cell in our data array and decide whether it is a land
point (and thus needs to be set to np.nan).
(For this example, we are going to define land as any grid point that is more than 50% land.)
PYTHON
ds = xr.open_dataset("data/pr_Amon_ACCESS-CM2_historical_r1i1p1f1_gn_201001-201412.nc")
clim = ds['pr'].mean("time", keep_attrs=True)
nlats, nlons = clim.data.shape
for y in range(nlats):
for x in range(nlons):
if sftlf.data[y, x] > 50:
clim.data[y, x] = np.nanWhile this approach technically works, the problem is that (a) the code is hard to read, and (b) in contrast to low level languages like Fortran and C, high level languages like Python and Matlab are built for usability (i.e. they make it easy to write concise, readable code) as opposed to speed. This particular array is so small that the looping isn’t noticably slow, but in general looping over every data point in an array should be avoided.
Fortunately, there are lots of numpy functions (which are written in
C under the hood) that allow you to get around this problem by applying
a particular operation to an entire array at once (which is known as a
vectorised operation). The np.where function, for instance,
allows you to make a true/false decision at each data point in the array
and then perform a different action depending on the answer.
The developers of xarray have built-in the np.where
functionality, so creating a new DataArray with the land masked becomes
a one-line command:
OUTPUT
<xarray.DataArray 'pr' (lat: 144, lon: 192)>
array([[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[7.5109556e-06, 7.4777777e-06, 7.4689174e-06, ..., 7.3359679e-06,
7.3987890e-06, 7.3978440e-06],
[7.1837171e-06, 7.1722038e-06, 7.1926393e-06, ..., 7.1552149e-06,
7.1576678e-06, 7.1592167e-06],
[7.0353467e-06, 7.0403985e-06, 7.0326828e-06, ..., 7.0392648e-06,
7.0387587e-06, 7.0304386e-06]], dtype=float32)
Coordinates:
* lon (lon) float64 0.9375 2.812 4.688 6.562 ... 353.4 355.3 357.2 359.1
* lat (lat) float64 -89.38 -88.12 -86.88 -85.62 ... 86.88 88.12 89.38
Attributes:
standard_name: precipitation_flux
long_name: Precipitation
units: kg m-2 s-1
comment: includes both liquid and solid phases
cell_methods: area: time: mean
cell_measures: area: areacellaMask option
Modify plot_precipitation_climatology.py so that the
user can choose to apply a mask via the following argparse
option:
PYTHON
parser.add_argument(
"--mask",
type=str,
nargs=2,
metavar=("SFTLF_FILE", "REALM"),
default=None,
help="""Provide sftlf file and realm to mask ('land' or 'ocean')""",
)This should involve defining a new function called
apply_mask(), in order to keep main() short
and readable.
Test to see if your mask worked by plotting the ACCESS-CM2 climatology for JJA:
BASH
$ python plot_precipitation_climatology.py data/pr_Amon_ACCESS-CM2_historical_r1i1p1f1_gn_201001-201412.nc JJA pr_Amon_ACCESS-CM2_historical_r1i1p1f1_gn_201001-201412-JJA-clim_land-mask.png --mask data/sftlf_fx_ACCESS-CM2_historical_r1i1p1f1_gn.nc ocean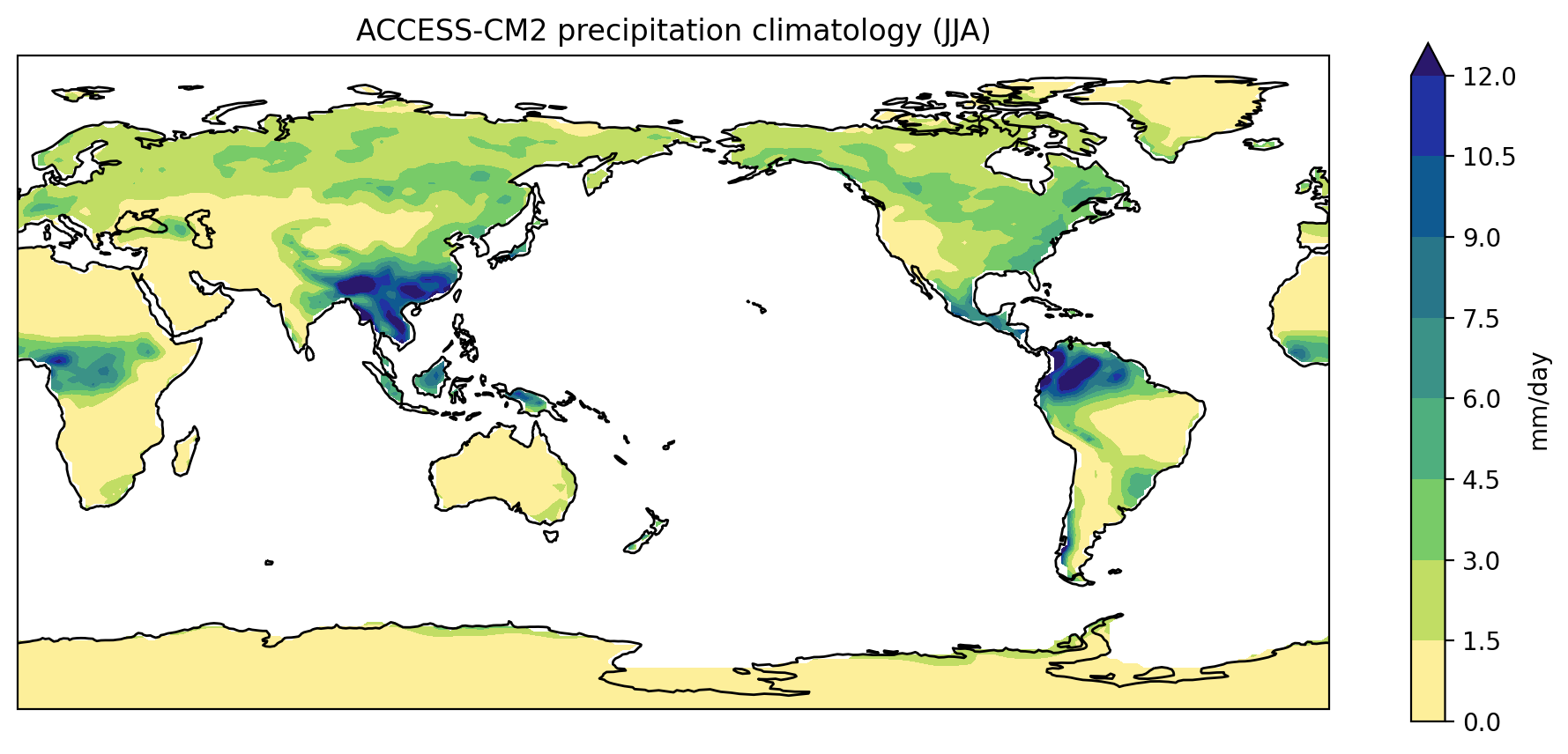
Commit the changes to git and then push to GitHub.
Make the following additions to
plot_precipitation_climatology.py (code omitted from this
abbreviated version of the script is denoted ...):
PYTHON
def apply_mask(da, sftlf_file, realm):
"""Mask ocean or land using a sftlf (land surface fraction) file.
Args:
da (xarray.DataArray): Data to mask
sftlf_file (str): Land surface fraction file
realm (str): Realm to mask
"""
ds = xr.open_dataset(sftlf_file)
if realm == 'land':
masked_da = da.where(ds['sftlf'].data < 50)
else:
masked_da = da.where(ds['sftlf'].data > 50)
return masked_da
...
def main(inargs):
"""Run the program."""
ds = xr.open_dataset(inargs.pr_file)
clim = ds["pr"].groupby("time.season").mean("time", keep_attrs=True)
clim = convert_pr_units(clim)
if inargs.mask:
sftlf_file, realm = inargs.mask
clim = apply_mask(clim, sftlf_file, realm)
...
if __name__ == "__main__":
description = "Plot the precipitation climatology for a given season."
parser = argparse.ArgumentParser(description=description)
...
parser.add_argument(
"--mask",
type=str,
nargs=2,
metavar=("SFTLF_FILE", "REALM"),
default=None,
help="""Provide sftlf file and realm to mask ('land' or 'ocean')""",
)
args = parser.parse_args()
main(args)plot_precipitation_climatology.py
At the conclusion of this lesson your
plot_precipitation_climatology.py script should look
something like the following:
PYTHON
import argparse
import xarray as xr
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
import numpy as np
import cmocean
def convert_pr_units(da):
"""Convert kg m-2 s-1 to mm day-1.
Args:
da (xarray.DataArray): Precipitation data
"""
da.data = da.data * 86400
da.attrs["units"] = "mm/day"
return da
def apply_mask(da, sftlf_file, realm):
"""Mask ocean or land using a sftlf (land surface fraction) file.
Args:
da (xarray.DataArray): Data to mask
sftlf_file (str): Land surface fraction file
realm (str): Realm to mask
"""
ds = xr.open_dataset(sftlf_file)
if realm == 'land':
masked_da = da.where(ds['sftlf'].data < 50)
else:
masked_da = da.where(ds['sftlf'].data > 50)
return masked_da
def create_plot(clim, model, season, gridlines=False, levels=None):
"""Plot the precipitation climatology.
Args:
clim (xarray.DataArray): Precipitation climatology data
model (str): Name of the climate model
season (str): Season
Kwargs:
gridlines (bool): Select whether to plot gridlines
levels (list): Tick marks on the colorbar
"""
if not levels:
levels = np.arange(0, 13.5, 1.5)
fig = plt.figure(figsize=[12,5])
ax = fig.add_subplot(111, projection=ccrs.PlateCarree(central_longitude=180))
clim.sel(season=season).plot.contourf(
ax=ax,
levels=levels,
extend="max",
transform=ccrs.PlateCarree(),
cbar_kwargs={"label": clim.units},
cmap=cmocean.cm.haline_r
)
ax.coastlines()
if gridlines:
plt.gca().gridlines()
title = f"{model} precipitation climatology ({season})"
plt.title(title)
def main(inargs):
"""Run the program."""
ds = xr.open_dataset(inargs.pr_file)
clim = ds["pr"].groupby("time.season").mean("time", keep_attrs=True)
clim = convert_pr_units(clim)
if inargs.mask:
sftlf_file, realm = inargs.mask
clim = apply_mask(clim, sftlf_file, realm)
create_plot(
clim,
ds.attrs["source_id"],
inargs.season,
gridlines=inargs.gridlines,
levels=inargs.cbar_levels
)
plt.savefig(
inargs.output_file,
dpi=200,
bbox_inches="tight",
facecolor="white",
)
if __name__ == "__main__":
description = "Plot the precipitation climatology for a given season."
parser = argparse.ArgumentParser(description=description)
parser.add_argument("pr_file", type=str, help="Precipitation data file")
parser.add_argument("season", type=str, help="Season to plot")
parser.add_argument("output_file", type=str, help="Output file name")
parser.add_argument(
"--gridlines",
action="store_true",
default=False,
help="Include gridlines on the plot",
)
parser.add_argument(
"--cbar_levels",
type=float,
nargs="*",
default=None,
help="list of levels / tick marks to appear on the colorbar",
)
parser.add_argument(
"--mask",
type=str,
nargs=2,
metavar=("SFTLF_FILE", "REALM"),
default=None,
help="""Provide sftlf file and realm to mask ('land' or 'ocean')""",
)
args = parser.parse_args()
main(args)Key Points
- For large arrays, looping over each element can be slow in high-level languages like Python.
- Vectorised operations can be used to avoid looping over array elements.
