Introduction to the dataset
Data
This workshop uses data from the enviromental experiment: Genomic adaptations in information processing underpin trophic strategy in a whole-ecosystem nutrient enrichment experiment, by Jordan G Okie et al. 2020 In this research, authors compared the differences between the microbial community in its natural, oligotrophic, phosphorus-deficient environment, a pond from the Cuatro Ciénegas Basin (CCB), and the same microbial community under a fertilization treatment.
All of the data used in this workshop can be downloaded from
Download data
The following commands download data from Zenodo into your computer. Type it in your command line.
wget https://zenodo.org/record/7010950/files/dc_workshop.zip?download=1
mv dc_workshop.zip?download=1 dc_workshop.zip
unzip dc_workshop.zip
Features of the dataset
The dataset in Zenodo contains two files:
dc_workshop.zipThree samples from Cuatro Ciénegas sediments with their corresponding taxonomic assignation files.MGRAST_MetaData_JP.xlsxMetadata about our three Cuatro Cienegas samples accepted by MGRAST
The dc_workshop.zip contains the following files.
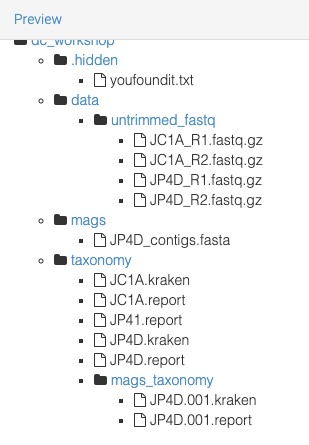
The directories are: .backup_dc_workshop,hidden, data, mags, and taxonomy.
Directory .backup_dc_workshop
Contains all the files produced or needed while runing the lesson.
Directory hidden
hidden contains a hidden file that will be used in the lesson Introduction to the Command Line for Metagenomics
episode 03 Navigating Files and Directories when learners will discover how to find hidden files.
Directory data
data contains four fastq files from two samples: JC1A and JP4D. These files
are the inputs of FastQC tool in the lesson Data Processing and Visualization for Metagenomics
next episodes two and three Assessing Read Quality and Trimming and Filtering.
In these episodes learners will remove bad quality nucleotides and prepare files for assembly and taxonomic assignation.
- JC1A_R1.fastq.gz 24.2 MB.
- JC1A_R2.fastq.gz 24.9 MB.
- JP4D_R1.fastq.gz 187.0 MB.
- JP4D_R2.fastq.gz 212.2 MB.
Directory mags
mags contains the assembly of the JP4D sample.
- JP4D_contigs.fasta 73.0 MB
Directory taxonomy
Since Kraken2 won’t be run in the lesson, this directory contains taxonomic
assignment obtained by running Kraken2 on the trimmed reads.
From these files users can obtained biom files that will be the input for
the R analysis and visualization of abundance.
Finally, it contains a subdirectory with the taxonomic assignment of the first bin from sample JP4D.
- JC1A.kraken 6.4 MB.
- JC1A.report 146.5 kB.
- JP41.report 375.9 kB.
- JP4D.kraken 74.8 MB.
- JP4D.report 404.2 kB
- mags_taxonomy
- JP4D.001.kraken 201.9 kB
- JP4D.001.report
References
Okie, J. G., Poret-Peterson, A. T., Lee, Z. M. P., Richter, A., Alcaraz, L. D., Eguiarte, L. E., Siefert, J. L., Souza, V., Dupont, C. L., & Elser, J. J. (2020). Genomic adaptations in information processing underpin trophic strategy in a whole-ecosystem nutrient enrichment experiment. ELife, 9. https://doi.org/10.7554/ELIFE.49816